AutoDock
| |||||||||||||||||||||||
Read other articles:
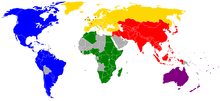
Bulu tangkis menjadi salah satu bagian dari ajang Pan American Games dimulai sejak tahun 1995 di Mar del Plata, Argentina. Kanada dan Amerika Serikat menjadi negara yang paling mendominasi pada even bulu tangkis sejak pertandingan ini dipertandingkan. Juara Sebelumnya Tahun Tunggal Putra Tunggal Putri Ganda Putra Ganda Putri Ganda Campuran 2007 Mike Beres Eva Lee Mike Beres William Milroy Eva Lee Mesinee Mangkalakiri Eva Lee Howard Bach 2003 Mike Beres Nigella Saunders Howard Bach Kevin Han C...

Cet article est une ébauche concernant une localité italienne et le Trentin-Haut-Adige. Vous pouvez partager vos connaissances en l’améliorant (comment ?) selon les recommandations des projets correspondants. Pour les articles homonymes, voir Castelnuovo. Castelnuovo Noms Nom allemand Neuburg Administration Pays Italie Région Trentin-Haut-Adige Province Trentin Code postal 38050 Code ISTAT 022049 Code cadastral C216 Préfixe tel. 0461 Démographie Population 1 08...

Surgical procedure of repairing corneal tissue to treat corneal blindness This article needs additional citations for verification. Please help improve this article by adding citations to reliable sources. Unsourced material may be challenged and removed.Find sources: Corneal transplantation – news · newspapers · books · scholar · JSTOR (July 2022) (Learn how and when to remove this template message) Corneal transplantationCornea transplant approximate...

Georgina Mace Georgina Mace lahir di London (Inggris) pada tahun 1953. Pada tahun 1976, ia memperoleh gelar sarjana bidang zoologi di Universitas Liverpool. Tiga tahun kemudian, ia dianugerahi gelar Ph.D di Universitas Sussex untuk studi ekologi evolusioner mamalia kecil. Mace kemudian meneliti dampak kawin sedarah pada populasi hewan di kebun binatang di Institusi Smithsonian Amerika Serikat. Pada tahun 2000, ia menjadi direktur ilmiah Institut Zoologi di London. Pada tahun 2006, ia diangkat...

Spanish novelist, poet, and playwright (1547–1616) Cervantes redirects here. For other uses, see Cervantes (disambiguation). Miguel Cervantes redirects here. For the American actor and singer, see Miguel Cervantes (actor). Miguel de CervantesThis portrait, attributed to Juan de Jáuregui,[a] is unauthenticated. No authenticated image of Cervantes exists.[1][2]Born29 September 1547 (assumed)Alcalá de Henares, Crown of CastileDied22 April 1616(1616-04-22) (aged 6...

County in Hebei, People's Republic of ChinaChangli County 昌黎县CountyChangliLocation in HebeiCoordinates: 39°43′N 119°10′E / 39.717°N 119.167°E / 39.717; 119.167CountryPeople's Republic of ChinaProvinceHebeiPrefecture-level cityQinhuangdaoCounty seatChangli Town (昌黎镇)Elevation22 m (72 ft)Population (2020 census)487,989[1]Time zoneUTC+8 (China Standard)Websitehttp://www.changli.gov.cn/ Changli (Chinese: 昌黎; pinyin: Ch...

Perotto (seduto a sinistra, in primo piano), con il team della P101 (eccetto Gaiti): De Sandre (seduto a destra), Garziera (in piedi a sinistra) e Toppi (in piedi a destra) Pier Giorgio Perotto (Torino, 24 dicembre 1930 – Genova, 23 gennaio 2002) è stato un ingegnere e informatico italiano. Progettista della Olivetti, fu un pioniere dell'informatica, noto soprattutto per aver progettato la Programma 101, il primo esempio di computer a programma memorizzato da tavolo[1][2] o...

2016年美國總統選舉 ← 2012 2016年11月8日 2020 → 538個選舉人團席位獲勝需270票民意調查投票率55.7%[1][2] ▲ 0.8 % 获提名人 唐納·川普 希拉莉·克林頓 政党 共和黨 民主党 家鄉州 紐約州 紐約州 竞选搭档 迈克·彭斯 蒂姆·凱恩 选举人票 304[3][4][註 1] 227[5] 胜出州/省 30 + 緬-2 20 + DC 民選得票 62,984,828[6] 65,853,514[6]...

Halaman ini berisi artikel tentang kawasan Polandia yang dianeksasi ke Jerman Nazi. Untuk kawasan lainnya yang diduduki pada 1939 namun tidak langsung dianeksasi, lihat Pemerintahan Umum. Kawasan Polandia yang dianeksasi oleh Jerman NaziPada warna tua, kawasan Polandia dianeksasi oleh Jerman Nazi dan Uni Soviet dengan Pemerintahan Umum semi-kolonial berwarna kuning muda (tengah)Pemisahan Polandia keempat – Pakta Nazi-Soviet Setelah Invasi Polandia pada permulaan Perang Dunia II, hampir sepe...

Siège de Boston Gravure représentant l'évacuation britannique de Boston. Informations générales Date 19 avril 1775 -17 mars 1776 Lieu Boston Issue Victoire américaine Belligérants Treize colonies Grande-Bretagne Commandants George Washington Thomas Gage Forces en présence 6 000 - 16 000 4 000 - 11 000 Pertes Environ 400 Environ 1 150 Guerre d'indépendance des États-UnisBatailles Campagne de Boston : Powder Alarm Suffolk Resolves Lexington et Concord Boston Thompson...

Scenic road in Colorado, United States Peak to Peak Scenic Byway National Forest Scenic BywayRoute informationMaintained by CDOTLength55 mi[1][2] (89 km)Existed1989–presentMajor junctionsSouth end SH 119 Black HawkNorth end US 34 / US 36 / SH 7 Estes Park LocationCountryUnited StatesStateColoradoCountiesGilpin, Boulder, Larimer counties Highway system Scenic Byways National National Forest BLM NPS Colorado...

3M 2013GénéralitésÉquipe Team 3MCode UCI MMMStatut Équipe continentalePays BelgiqueSport Cyclisme sur routeEffectif 17PalmarèsNombre de victoires 03M 2014modifier - modifier le code - modifier Wikidata La saison 2013 de l'équipe cycliste 3M est la première de cette équipe. Préparation de la saison 2013 Sponsors et financement de l'équipe Cette section est vide, insuffisamment détaillée ou incomplète. Votre aide est la bienvenue ! Comment faire ? Arrivées et dép...

لمعانٍ أخرى، طالع كارل ريتر (توضيح). كارل ريتر معلومات شخصية الميلاد 7 نوفمبر 1888 [1] فورتسبورغ الوفاة 7 أبريل 1977 (88 سنة) [1] بوينس آيرس مواطنة ألمانيا الحياة العملية المهنة مخرج أفلام، وكاتب سيناريو، ومنتج أفلام، ورسام توضيحي الح�...

Air Education and Training Command Création 1er juillet 1993 Pays États-Unis Branche United States Air Force Type Commandement majeur Effectif 62 000 Garnison Randolph Air Force Base, Texas Surnom AETC Commandant Stephen R. Lorenz modifier L'Air Education and Training Command (AETC) est l'un des grands commandements de l'USAF. Il est formé le 1er juillet 1993, des suites de la fusion de l'Air Training Command et de l'Air University. Son quartier général est s...

American judge (1861–1927) John Carter RoseJudge of the United States Court of Appeals for the Fourth CircuitIn officeDecember 20, 1922 – March 26, 1927Appointed byWarren G. HardingPreceded bySeat established by 42 Stat. 837Succeeded byElliott NorthcottJudge of the United States District Court for the District of MarylandIn officeApril 4, 1910 – December 26, 1922Appointed byWilliam Howard TaftPreceded bySeat established by 36 Stat. 201Succeeded byMorris Ames SoperUnite...

The Battle of the Arar was fought between the migrating tribes of the Helvetii and six Roman legions under the command of Gaius Julius Caesar in 58 BC. It was the first major battle of the Gallic Wars and ended in a tactical victory for the outnumbered Roman army. This article needs additional citations for verification. Please help improve this article by adding citations to reliable sources. Unsourced material may be challenged and removed.Find sources: Battle of the Arar – ...

Carl Jung's Liber Novus (Red Book), and Psychology and Alchemy. This is a list of writings published by Carl Jung. Many of Jung's most important works have been collected, translated, and published in a 20-volume set by Princeton University Press, entitled The Collected Works of C. G. Jung. Works here are arranged by original publication date if known. N.B. See also Juliette Vieljeux, Catalogue chronologique des écrits de Carl Gustav Jung, Cahiers Jungiens de Psychanalyse, hors-série, 1996...

صورة لتشريح أذن إنسان، بقرة، كلب، حصان، نمر، قط، وفئران، خنزير وخروف واوز هو موضح للعالم والراهب الألماني أثانيسيوس كيرتشر. العلاقة بين المسيحية والطب تعود إلى عصور المسيحية المبكرة إذ كان للمفاهيم المسيحية من الرعاية ومساعدة المرضى دور في تطوير الأخلاق الطبية.[1] أنش...

British princess; eldest daughter of Frederick, Prince of Wales For her niece Augusta Sophia (1768–1840), see Princess Augusta Sophia of the United Kingdom. Princess AugustaPortrait by Johann Georg Ziesenis, c. 1764-70Duchess consort of Brunswick-LüneburgPrincess consort of Brunswick-WolfenbüttelTenure26 March 1780 – 10 November 1806Born(1737-07-31)31 July 1737St James's Palace, LondonDied23 March 1813(1813-03-23) (aged 75)Hanover Square, LondonBurial31 March 1813Royal Vault,...

16th-century English nobleman The Right HonourableThe Earl of SurreyKGHenry Howard, Earl of Surrey. Painting in the Mannerist style possibly attributed to the Dutch artist William Scrots. c. 1546.Personal detailsBornc. 1517Hunsdon, HertfordshireDied19 January 1547 (aged 29 or 30)Tower Hill, London, EnglandResting placeFirst at the Church of All Hallows, Tower Street, London and then at Church of St Michael the Archangel, Framlingham, SuffolkSpouseFrances de VereChildrenThomas Howard, 4th Duke...
