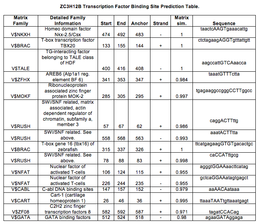
ZC3H12B
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Read other articles:

Provinsi Ise (伊勢国code: ja is deprecated , ise no kuni) atau dikenal sebagai Seishu (勢州code: ja is deprecated , seishū) adalah nama provinsi lama Jepang yang menempati hampir seluruh wilayah prefektur Mie sekarang. Ise berbatasan dengan provinsi Iga, Kii, Mino, Omi, Owari, Shima, dan provinsi Yamato. Ibu kota berada di Suzuka. Kota istana yang terbesar adalah kota yang sekarang dikenal sebagai Tsu, walaupun ada juga kota sekeliling istana yang berkembang di tempat-tempat yang memil...

Public house in Marylebone, London The Beehive, MaryleboneThe Beehive pub at 126 Crawford StreetTypePublic houseLocationMaryleboneCoordinates51°31′12″N 0°09′26″W / 51.5200°N 0.1573°W / 51.5200; -0.1573OS grid referenceTQ 27944 81734AreaCity of WestminsterBuiltc. 1793Rebuilt1884 Listed Building – Grade IIOfficial nameThe Beehive public houseDesignated1 December 1987Reference no.1221026 Location of The Beehive, Marylebone in Central London The Beehive ...

North, Central America and Caribbean Volleyball ConfederationOlahragaBola voli Voli pantaiYurisdiksiAmerika Tengah dan UtaraJumlah anggota42 negaraSingkatanNORCECABerdiri1968AfiliasiFIVBKantor pusatSanto Domingo,Republik DominikaPresiden Cristóbal Marte HoffizSitus web resmiwww.norceca.org North, Central America and Caribbean Volleyball Confederation (NORCECA) (Indonesia: Konfederasi Bola Voli Amerika Utara dan Tengah) adalah induk organisasi kontinental untuk olahraga bola voli di Amerika U...

Tschammerpokal 1935DFB Pokal 1935 Competizione DFB-Pokal Sport Calcio Edizione 1ª Date dal 6 gennaio 1935all'8 dicembre 1935 Luogo Germania Risultati Vincitore Norimberga(1º titolo) Secondo Schalke 04 Cronologia della competizione 1936 Manuale La Tschammerpokal del 1935 fu la prima edizione della coppa nazionale della Germania. Il nome fu scelto in onore del Reichssportführer Hans von Tschammer und Osten, il più alto responsabile dello sport nel Terzo Reich. Iniziò...

Questa voce sull'argomento stagioni delle società calcistiche italiane è solo un abbozzo. Contribuisci a migliorarla secondo le convenzioni di Wikipedia. Segui i suggerimenti del progetto di riferimento. Voce principale: Circolo Sportivo Trevigliese. Circolo Sportivo TreviglieseStagione 1923-1924Sport calcio Squadra Trevigliese Seconda Divisione4º posto nel girone D. StadioCampo C.S. Trevigliese 1922-1923 1924-1925 Si invita a seguire il modello di voce Questa pagina raccoglie l...

Chinese painter (713–755) Not to be confused with Zheng Xuan. For Taiwanese singer, see Deserts Chang. Spring Outing of the Tang Court, by Zhang Xuan. Zhang Xuan (traditional Chinese: 張萱; simplified Chinese: 张萱; pinyin: Zhāng Xuān; Wade–Giles: Chang Hsüan) (713–755) was a Chinese painter who lived during the Tang dynasty (618–907). Zhang Xuan painted many pieces of art, one of his best-known paintings is Court Ladies Preparing Newly Woven Silk, of which a si...

NCSIST Albatross Role UAVType of aircraft Manufacturer National Chung-Shan Institute of Science and Technology Introduction 2007 Status In service Primary user Republic of China Navy Number built >32 Developed from Chung Shyang I NCSIST Albatross, then called the Chung Shyang II UAV, being unveiled publicly in 2007 UAV 9717 on display at CKS Memorial Hall UAV 9717 on display at No.11 Pier Covered UAV 9728 at Chengkungling Ground with rain cover The Albatross(Chinese: 銳鳶; piny...

阿尔弗雷德·金赛1955年11月在法蘭克福的金賽出生阿爾弗雷德·查爾斯·金賽1894年6月23日[1] 美國新泽西州霍博肯[1]逝世1956年8月25日(1956歲—08—25)(62歲) 美國印第安納州布卢明顿[1]国籍 美國母校史蒂文斯理工學院鲍登学院哈佛大学知名于針對人類的性學研究:金赛报告、金賽性、性別與生殖研究中心、金賽量表科学生涯研究领域生物学机构印第...

Частина серії проФілософіяLeft to right: Plato, Kant, Nietzsche, Buddha, Confucius, AverroesПлатонКантНіцшеБуддаКонфуційАверроес Філософи Епістемологи Естетики Етики Логіки Метафізики Соціально-політичні філософи Традиції Аналітична Арістотелівська Африканська Близькосхідна іранська Буддій�...

Croatian food company, known worldwide mostly for producing Vegeta Podravka d.d.Official logoCompany typePublic companyTraded asZSE: PODRISINHRPODRRA0004 IndustryFood and beveragesFounded1934; 90 years ago (1934)FounderMarijan & Matija WolfHeadquartersKoprivnica, CroatiaArea servedWorldwideKey peopleMartina Dalić, ph.d. (Chairman)[1] Željko Vukina (President of the Supervisory Board)[2]ProductsFood seasoning, soups, ready-to-serve meals, half-...

This article relies largely or entirely on a single source. Relevant discussion may be found on the talk page. Please help improve this article by introducing citations to additional sources.Find sources: Herrerism – news · newspapers · books · scholar · JSTOR (February 2017) Political party in Uruguay Herrerism LeaderLuis Lacalle PouIdeologyLiberal conservatismPolitical positionCentre-rightNational affiliationNational PartyPolitics of Uruguay...

Sint Maarten politician William Marlin3rd Prime Minister of Sint MaartenIn office19 November 2015 – 24 November 2017MonarchWillem-AlexanderGovernorEugene HolidayPreceded byMarcel GumbsSucceeded byRafael Boasman Personal detailsBorn (1950-10-21) 21 October 1950 (age 73)Sint MaartenPolitical partyNational Alliance William Marlin (born 21 October 1950)[1] is a Sint Maarten politician who served as the 3rd Prime Minister of Sint Maarten from 2015 to 2017.[2][3&...

Indian businessman Russi ModiBorn17 January 1918Bombay, Bombay Presidency, British IndiaDied16 May 2014(2014-05-16) (aged 96)Kolkata, IndiaAlma materOxfordOccupationChairman of Tata Steel from 1984 to 1993SpouseSiloo Russi ModyParent(s)Sir Homi Mody and Lady Jerbhai Rustomji Homusji Mody,[1] known to most as Russi Mody (17 January 1918 – 16 May 2014), was a chairman and managing director of Tata Steel and a leading member of the Tata Group. Early years Russi was born ...

American sports pay television network For the Canadian version of this channel, see NBA TV Canada. For the Philippine version of this channel, see NBA TV Philippines. Television channel NBA TVCountryUnited StatesBroadcast areaNationwideHeadquartersAtlanta, Georgia, U.S.ProgrammingLanguage(s)EnglishPicture format1080i HDTV(downscaled to letterboxed 480i for the SDTV feed)OwnershipOwnerNational Basketball Association(operated by TNT Sports)Sister channelsMLB NetworkMotor TrendHistoryLaunchedNo...
Phoenix RacewayPhoenix Raceway as of 2019Lokasi7602 S Avondale BoulevardAvondale, Arizona 85323United StatesZona waktuUTC−7Kapasitas42,000[1]PemilikNASCARDibuka1964Nama sebelumnyaPhoenix International Raceway (1964–1973, 1976–2017)FasTrack International Speedway (January 1973–August 1976) Jeff Gordon Raceway (November 15, 2015)ISM Raceway (2018–January 2020)Acara besarNASCAR Cup SeriesNASCAR Xfinity SeriesNASCAR Camping World Truck SeriesDogleg ovalPermukaanAsphaltPanjang1.0...

Cet article est une ébauche concernant une localité allemande. Vous pouvez partager vos connaissances en l’améliorant (comment ?) selon les recommandations des projets correspondants. Dohr Armoiries Administration Pays Allemagne Land Rhénanie-Palatinat Arrondissement(Landkreis) Cochem-Zell Bourgmestre(Ortsbürgermeister) Alois Franzen Code postal 56812 Code communal(Gemeindeschlüssel) 07 1 35 021 Indicatif téléphonique 02671 Immatriculation COC Démographie Population 625 ...

National park in Kırklareli, Turkey İğneada Floodplain Forests National Parkİğneada Longoz Ormanları Milli Parkıİğneada Floodplain Forests NPLocationKırklareli Province, TurkeyNearest cityİğneada, DemirköyCoordinates41°49′10″N 27°57′14″E / 41.81944°N 27.95389°E / 41.81944; 27.95389Area3,155 ha (7,800 acres)EstablishedNovember 13, 2007; 16 years ago (2007-11-13)Governing bodyDirectorate-General of Nature Protecti...

Cup-style tournament US Arena Open CupSportIndoor soccerFounded2008Most recentchampion(s)Metro Louisville FC (2021-22)Most titlesSan Diego Sockers (3)RelatedcompetitionsMASL, PASLOfficial websiteU.S. Open Championship The United States Open for Arena Soccer is a cup-style tournament for all Major Arena Soccer League, MASL2, MASL3 and Premier Arena Soccer League teams. Established in 2008, the PASL announced they would hold the first tournament for indoor soccer open to all leagues and/or exis...

Alunda moose at the Swedish History Museum The Alunda moose or the Alunda axe is a Neolithic stone axe c.2000 B.C.that was found during the excavation of a ditch in Norrlövsta i Alunda parish, Östhammar Municipality, Uppland in 1920.[1] The Alunda moose is considered one of the most beautiful animal sculptures of the Nordic neolithic and is a 21 cm. ceremonial axe carved from greenish black diorite. One side is shaped like a moose head. The sculpture was realized in a naturalistic a...

Pour les articles homonymes, voir Gouraud et Henri Gouraud. Si ce bandeau n'est plus pertinent, retirez-le. Cliquez ici pour en savoir plus. Certaines informations figurant dans cet article ou cette section devraient être mieux reliées aux sources mentionnées dans les sections « Bibliographie », « Sources » ou « Liens externes » (décembre 2023). Vous pouvez améliorer la vérifiabilité en associant ces informations à des références à l'aide d'appe...